Aik Choon Tan, Daniel Q. Naiman, Lei Xu, Raimond L. Winslow and Donald Geman
Simple decision rules for classifying human cancers from gene expression profiles
Bioinformatics 2005 21(20):3896-3904
[PDF][WebSite]
・サンプル分類法として、k-TSP (k-Top Scoring Pairs) を提案する。これは過去提案されたTSP [Geman,2004] の改良版である。
・データ
#Binary class
1.Colon [Alon]
2.Leukemia [Golub]
3.CNS [Pomeroy]
4.DLBCL [Shipp]
5.Lung [Gordon]
6.Prostate1 [Singh]
7.Prostate2 [Stuart]
8.Prostate3 [Welsh]
9.GCM [Ramaswamy]
#Multi-class
1.Leukemia1 [Golub]
2.Lung1 [Beer]
3.Luekemia2 [Armstrong]
4.SRBCT [Khan]
5.Breast [Perou]
6.Lung2 [Bhattacharjee]
7.DLBCL [Alizadeh]
8.Leukemia3 [Yeoh]
9.Cancers [Su]
10.GCM [Ramaswamy]
・比較法
1.TSP
2.k-TSP
3.DT (C4.5 decision tree)
4.NB (Naive Bayes)
5.k-NN (k-nearest neighbor)
6.SVM (Support Vector Machines)
7.PAM (Prediction analysis of microarrays)
・クラス分けの評価法
#Binary class:識別率はLOOCVで計算
#Multi class:実験の設定は以下の三つ
1.One-vs-Others (1-vs-r)
2.One-vs-One (1-vs-1)
3.Hierarchical Classification (HC)
・問題点「Current methods generate classifiers that are accurate but difficult to interpret. This is the trade-off between credibility and comprehensibility of the classifiers.」
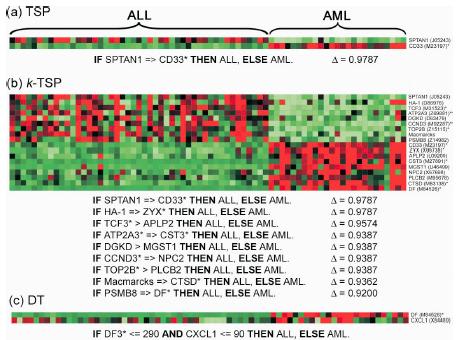
・k-TSPとは「k-TSP, a refinement of the original TSP algorithm, which uses exactly k pairs of genes for classifying gene expression data. When k = 1, this algorithm, referred to simply as TSP necessarily selects a unique pair of genes. More generally, both TSP and k-TSP may be seen as special cases of a new classification methodology based on the concept of ‘relative expression reversals.'」
・「This is accomplished by basing the classification on the k disjoint Top Scoring Pairs (k-TSP) of genes that achieve the best combined score.」
Simple decision rules for classifying human cancers from gene expression profiles
Bioinformatics 2005 21(20):3896-3904
[PDF][WebSite]
・サンプル分類法として、k-TSP (k-Top Scoring Pairs) を提案する。これは過去提案されたTSP [Geman,2004] の改良版である。
・データ
#Binary class
1.Colon [Alon]
2.Leukemia [Golub]
3.CNS [Pomeroy]
4.DLBCL [Shipp]
5.Lung [Gordon]
6.Prostate1 [Singh]
7.Prostate2 [Stuart]
8.Prostate3 [Welsh]
9.GCM [Ramaswamy]
#Multi-class
1.Leukemia1 [Golub]
2.Lung1 [Beer]
3.Luekemia2 [Armstrong]
4.SRBCT [Khan]
5.Breast [Perou]
6.Lung2 [Bhattacharjee]
7.DLBCL [Alizadeh]
8.Leukemia3 [Yeoh]
9.Cancers [Su]
10.GCM [Ramaswamy]
・比較法
1.TSP
2.k-TSP
3.DT (C4.5 decision tree)
4.NB (Naive Bayes)
5.k-NN (k-nearest neighbor)
6.SVM (Support Vector Machines)
7.PAM (Prediction analysis of microarrays)
・クラス分けの評価法
#Binary class:識別率はLOOCVで計算
#Multi class:実験の設定は以下の三つ
1.One-vs-Others (1-vs-r)
2.One-vs-One (1-vs-1)
3.Hierarchical Classification (HC)
・問題点「Current methods generate classifiers that are accurate but difficult to interpret. This is the trade-off between credibility and comprehensibility of the classifiers.」
・k-TSPとは「k-TSP, a refinement of the original TSP algorithm, which uses exactly k pairs of genes for classifying gene expression data. When k = 1, this algorithm, referred to simply as TSP necessarily selects a unique pair of genes. More generally, both TSP and k-TSP may be seen as special cases of a new classification methodology based on the concept of ‘relative expression reversals.'」
・「This is accomplished by basing the classification on the k disjoint Top Scoring Pairs (k-TSP) of genes that achieve the best combined score.」















※コメント投稿者のブログIDはブログ作成者のみに通知されます